Artwork
This illustration depicts the molecular structure of an HIV drug known as an INSTI (at center) bound to the HIV intasome active site. The intasome is the viral machine that allows HIV to infect human immune cells. By filling the binding pocket of the intasome, this drug blocks the normal function of the intasome, thereby preventing the virus from establishing an infection in the target cell.
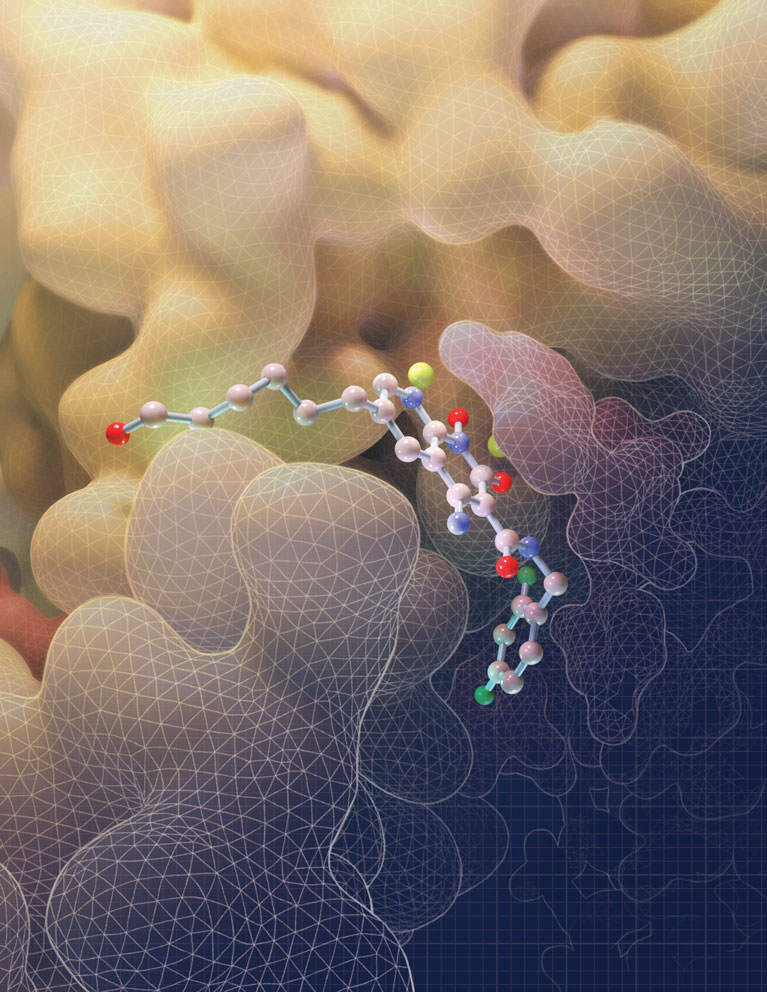
Image was created by Amy Cao. Work was published in: Passos, D.O., Li, M., Jó?wik, I.K., Zhao, X.Z., Santos-Martins, D., Yang, R., Smith, S.J., Jeon, Y., Forli, S., Hughes, S.H., Burke, T.R., Craigie, R., Lyumkis, D. Structural basis for strand transfer inhibitor binding to HIV intasomes. (2020) Science. DOI: 10.1126/science.aay8015
Whereas single-particle cryo-EM is a rapidly advancing technique, resolution anisotropy caused by “preferred specimen orientation” has remained a stifling problem for many important protein samples. The preferred orientation problem refers to the preference of proteins to “stick” to a surface on a cryo-EM grid in one or several discreet orientations, thus limiting the number of angular views that can be observed of the protein of interest. Researchers have described a generalizable strategy to tackle the problem of preferred specimen orientation by tilting the sample inside the microscope and collecting data at different angles. Just as looking at soup cans from different angles lets you see different shapes, viewing proteins at a tilt reveals different aspects of their structure and improves overall resolution.

The image was created by Jamie Simon, Dmitry Lyumkis, and Yong Zi Tan. The work was published in:
Yong Zi Tan, Philip R Baldwin, Joseph H Davis, James R Williamson, Clinton S Potter, Bridget Carragher, Dmitry Lyumkis. (2017) Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods, doi:10.1038/nmeth.4347
Structure of a complex between HIV-1 Integrase with viral and target (host) DNA, collectively called an intasome. Intasomes mediate the irreversible integration of retroviral DNA into target host cells, and the HIV intasome is targeted by some of the best available antiretroviral therapeutics. An atomic understanding of the HIV intasome structure should facilitate efforts to develop better therapeutics with improved resistance profiles.
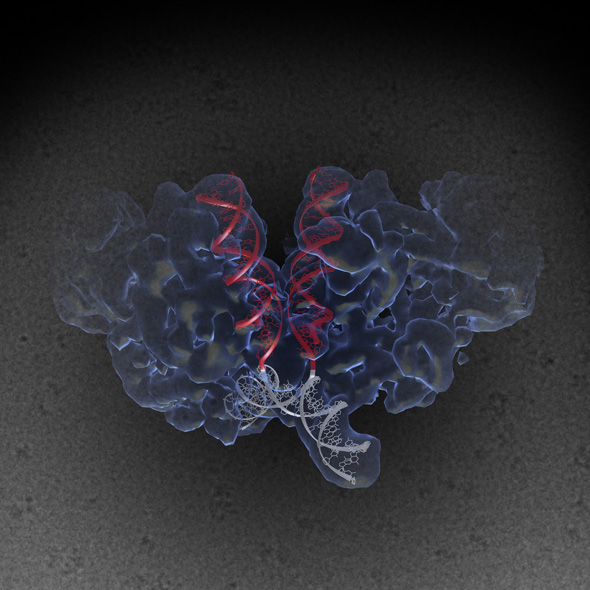
The image was created by Jamie Simon and Dario Passos. Work that led to the 3D map was published in:
Passos D., Li M., Yang R., Rebensburg S., Ghirlando R., Jeon Y., Shkriabai N., Kvaratskhelia M., Craigie R., Lyumkis D. (2017) CryoEM Structures and Atomic Models of the HIV-1 Strand Transfer Complex Intasome. Science, 355(6320), 89-92
Salk researchers captured the structure of a protein complex called an intasome (center) that lets viruses similar to HIV establish permanent infection in their hosts. The intasome hijacks host genomic material, DNA (white) and histones (beige), and irreversibly inserts viral DNA (blue).
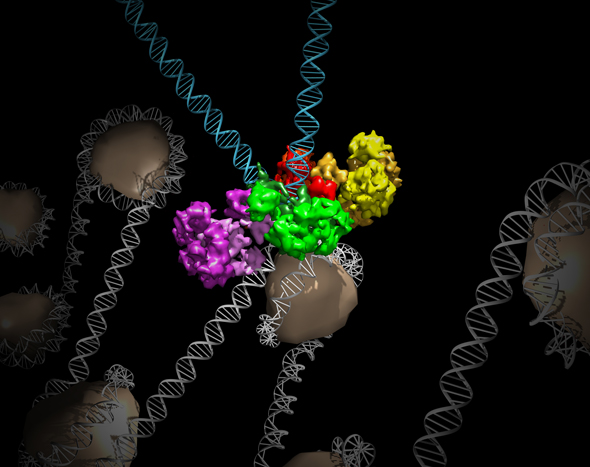
The image was created by Jamie Simon and Dmitry Lyumkis. Work that led to the 3D map was published in:
Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, & Engelman AN. (2016). Cryo-EM reveals a novel octameric integrase structure for ?-retroviral intasome function. Nature, 530(7590), 358—361.
Structures of the HIV Envelope trimer, a key AIDS molecule, modeled onto a viral membrane. These are the first atomic-level structures of the tripartite HIV molecule — long considered one of the most difficult targets in structural biology and of great value for medical science. New data provide the most detailed picture yet of the AIDS-causing virus’s complex Envelope structure, including sites that future vaccines will try to mimic to elicit a protective human immune response. The structures were solved independently by x-ray crystallography and cryo-electron microscopy.
The image was rendered by Graham Johnson (UCSF). Work that led to the 3D map was published in:
Lyumkis D, Julien JP, de Val N, Cupo A, PoUer CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013; 342:1484-90.
Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013; 342:1477-83.
Artist’s rendition of recent advances in single-particle electron microscopy. The advent of cameras that are capable of recording “movies” rather than individual images, provides the microscopist with the ability to deblur motion captured within an electron micrograph. This ability leads to higher resolution within resulting 3D reconstructions of the imaged object, ultimately enabling one to address increasingly complicated questions about the macromolecular assembly.

The image was rendered by Arne Moeller (formerly TSRI, now at Aarhus University). Work that led to the 3D map was published in:
Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, Grigorieff N. (2012). Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure, 20, 1823-1828.
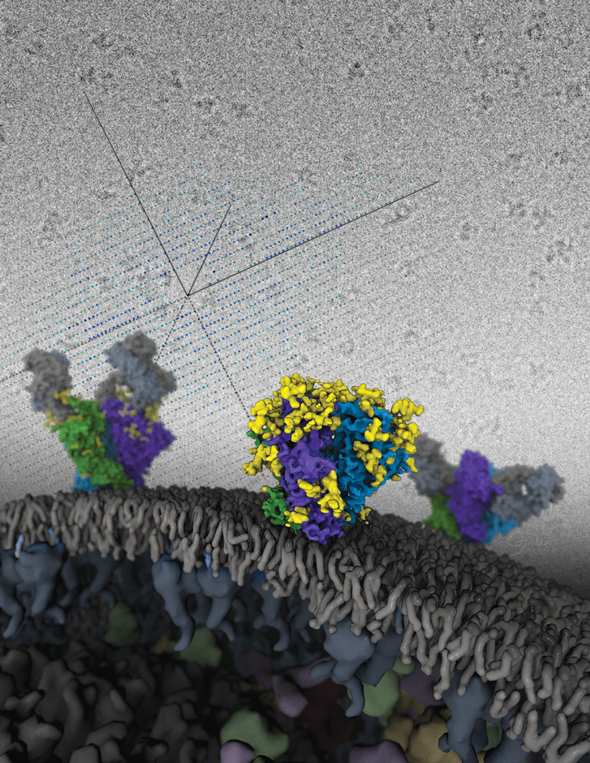
Cryo-electron microscopy, in combination with other biophysical techniques, was used to characterize the macromolecular composition, structural details, and allosteric requirements of the SgrAI restriction endonuclease. The results showed, in response to activation brought upon by invading phage, the SgrAI restriction enzyme forms run-on oligomers that are allosterically modulated by viral DNA. The enzyme’s mechanism of activation appears to be unique within the enzymatic world.
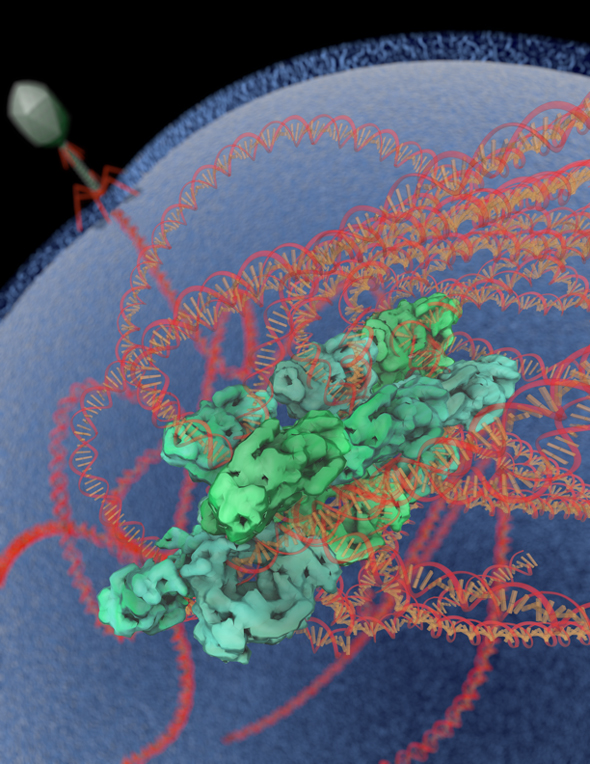
The image was rendered by Arne Moeller (formerly TSRI, now at Aarhus University). Work that led to the 3D map was published in:
Lyumkis D, Talley H, Stewart A, Shah S, Park CK, Tama F, Potter CS, Carragher B, Horton NC. (2013) Allosteric regulation of DNA cleavage and sequence-specificity through run-on oligomerization. Structure, 21, 1-11.